Due to the dynamic nature of this research project we’d like to summarize the many directions of this project and give closure to the work that did not end up in the Executive Summary Post.
Defining Structure-Relevant Statistics
To define spacial correlations both 2pt statistics and Chord Length Distribution were collected for both structures. The 2pt statistics were collected using non-periodic conditions, and truncated to capture maximum 100-pixel autocorrelations. With this truncation the number of statistics for each structure was 7,880,599 compared to the maximum 900 for CLDs. Our original workflow is shown below:
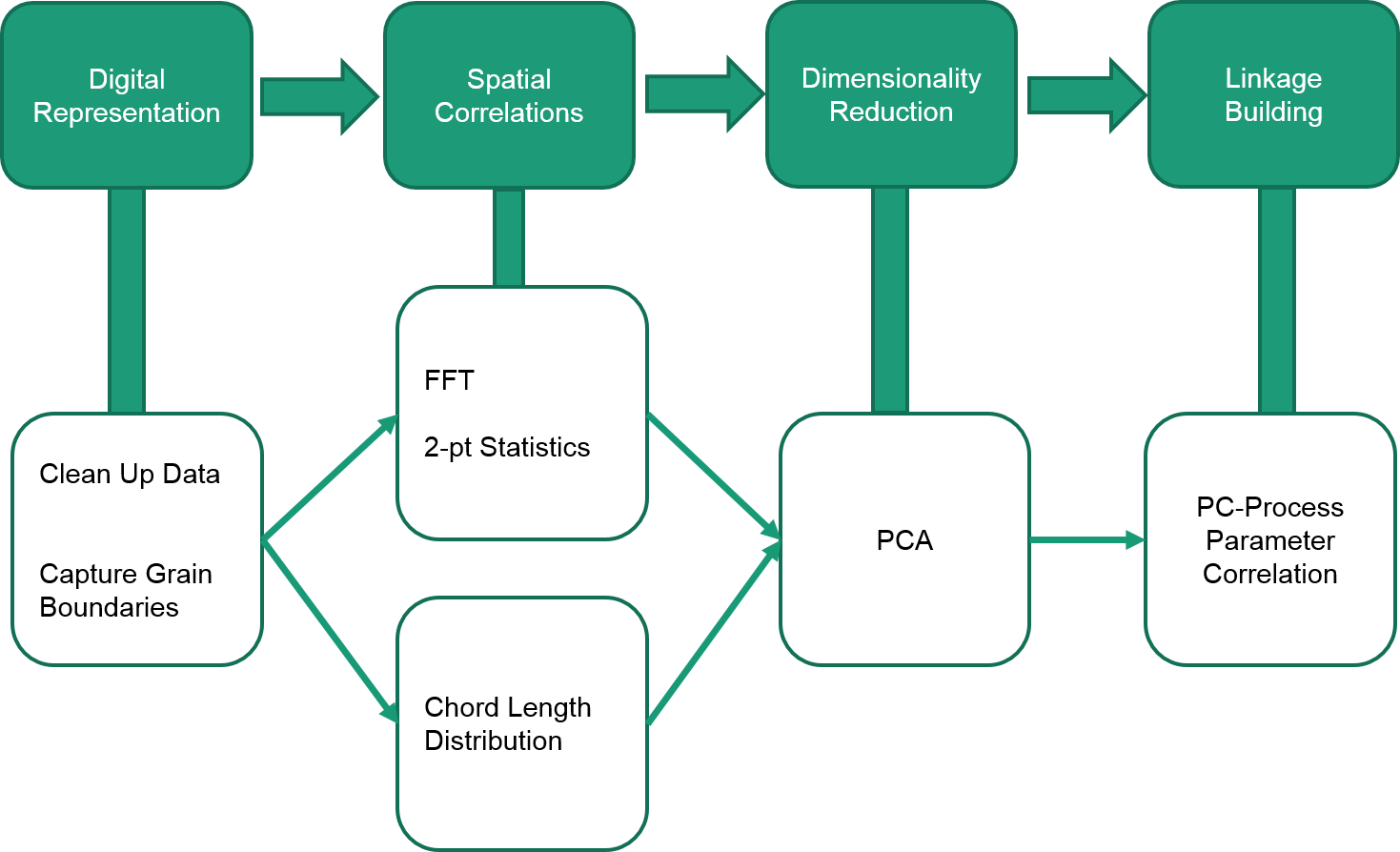 Fig.1. Original workflow
Fig.1. Original workflow
The CLD statistics presented no problem to analyze and we used them to to PCA and build Process-Structure Linkages using Linear Polynomial Regression. However the 2pt statistics became too unwieldy to analyze due to their size. The microstructure ensemble represented by 2pt statistics was about 90GB memory and over 260GB for PCA algorithm to run. Our access to high RAM computing nodes was limited and the set computation could not be run. The PCA of small subsets of our dataset showed promising separation in PC space and future work will include 2pt statistics analysis given the necessary tools. This may lend us improved linkages in certain PC dimensions.
Reconsidering Truncation of the Dataset
The 2pt statistics PCA could also give us access to the ‘low-confidence’ structures because the statistic is well defined within the simulation space for all structures. This would enable us to extend the model prediction range rather than truncating the dataset to exclude these structures.