This is the post equivalent of the presentation given for our second progress report.
Outline
- Detailed Workflow and Data Pipeline
- Conditioning and Segmentation in Matlab (Step 1)
- 2 Point Statistics and PCA in Python (Steps 2, 3)
- Preliminary Results
Matlab
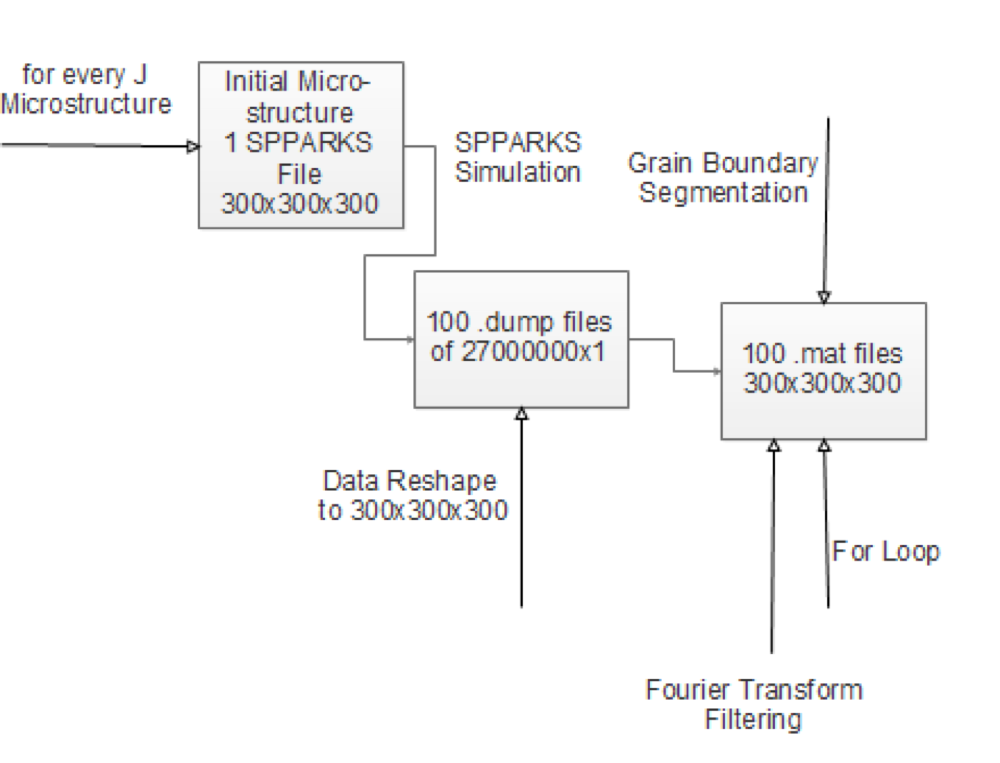
Image Segmentation
- Output from SPPARKS is a 27 million by one vector, reshape it to a 300X300X300 matrix.
- Each number corresponds to a grainID. Max value corresponds to a precipitate.
- We need to segment this data into our Microstructure Function.
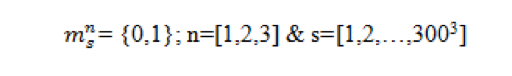
Obtaining Grain Boundary and Precipitates
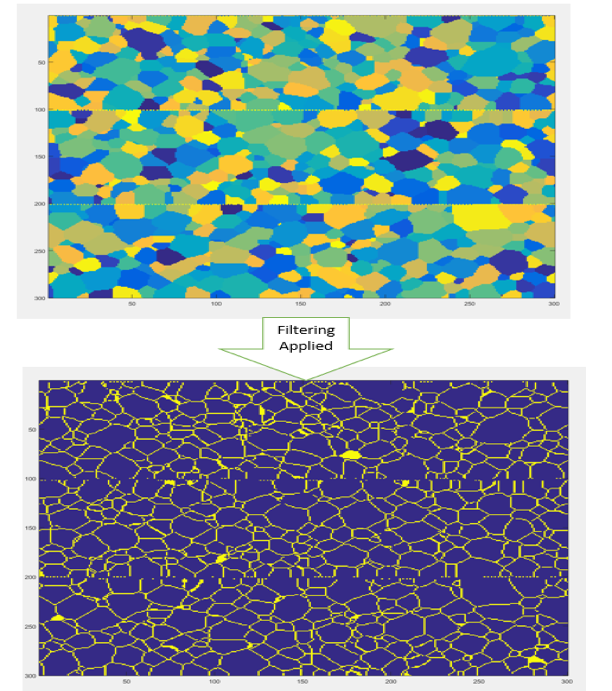
Obtaining .mat files using PACE
- The issue is that the output from the filter are 3 images with 0 and 1.
- Which is desirable but we need it to change to one image with 3 distinct phases for us to input it to PyMKS.
- Made a for loop that will go through each
.dumpfileand automatically segment it and save it as .mat file.
Python
Continuing where we left off in Matlab…
- Input:
.matfiles that only contain 0, 1, and 2 as values in an 300x300x300 array- Greatly reduces memory:
file.dump(~500MB) ->file.mat(~3MB)
2 Point Statistics Conditioning
- Initial size: (300, 300, 300) per file
- Loop that pulls each 100 outputs per simulation
- Concatenates to feed directly into PyMKS
- (n_samples, x, y, z) = (100, 300, 300, 300) per simulation
2 Point Statistics Computation
- 3 phase material -> 6 correlations
- Results in array of size (100, 300, 300, 300, 6)
- Initially only care about one correlation: grain boundary and pins [1,2]
- After 2 point statistics computation: (100, 300, 300, 300, 1)
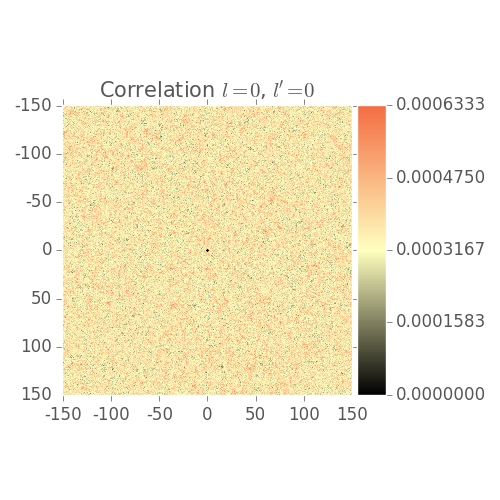
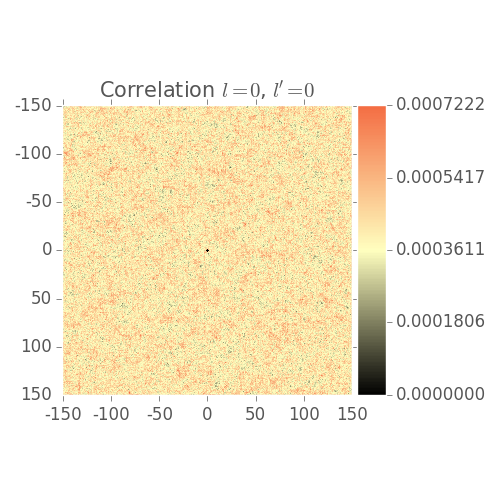
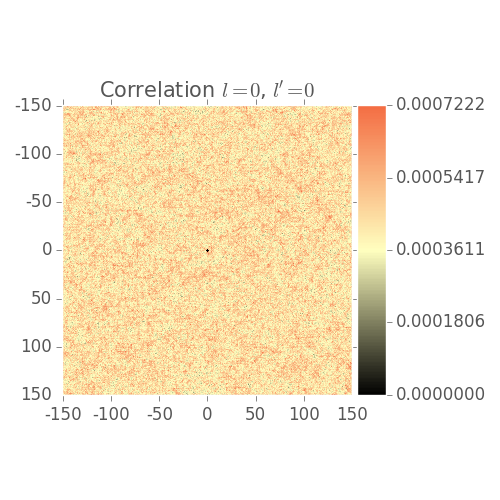
2 Point Statistics Plots
Shows 2 point statistics evolve over grain growth simulation
Note: Visualization issues:
- Low volume fraction
- Color bar not scaled correctly by default
PCA
- Forming the Data Matrix
- Consider each 3D microstructure as long array
- Reshape (100, 300, 300, 300) to (100, 300^3) = (100, 27,000,000)
- SciKit Learn Randomized PCA
- Can specify number of PC values to output
- Have tried: 2 and 3
- Results in (100, 2) and (100, 3) matrix
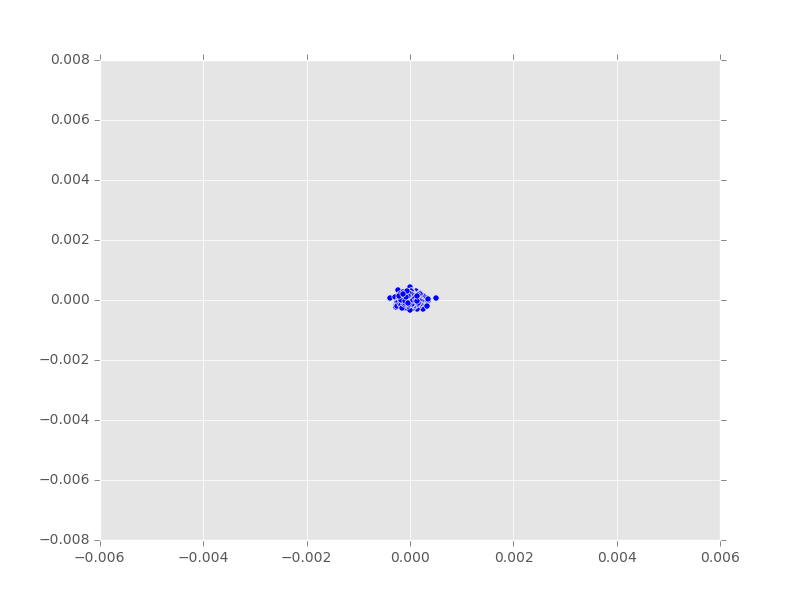
PCA Plots
- Final
.matfiles currently finishing on PACE - Code pipeline is working but needs polishing
- Meaningless 2D and 3D example below
Next Steps
Bring analysis and visualization into 3D.
- 2 point statistics of 3D data (longer computation)
- Visualize 3D 2 point statistics results
- Compute 3D PCA and Plot to see a path per simulation
- Work with David Brough to use latest fitting code to create model